nf-core/magmap
Best-practice analysis pipeline for mapping reads to a (large) collections of genomes
Introduction
nf-core/magmap is a bioinformatics best-practice analysis pipeline that maps reads to (large) collections of genomes. Its main output are tables with quantification of features (genes) in genomes which can be analyzed in R, Python or by other pipelines such as nf-core/differentialabundance. It is mainly meant for metatranscriptomes and metagenomes, but can be used for other types of samples where mapping to contigs is relevant. The nf-core/rnaseq pipeline is similar in purpose, but meant for single organisms with reference genomes and annotations, in practice eukaryotic model organisms.
Before running this pipeline, consider running nf-core/taxprofiler to get a view of whether the community composition in your samples fit nf-core/magmap and its focus on prokaryotic genomes.
The pipeline is built using Nextflow, a workflow tool to run tasks across multiple compute infrastructures in a very portable manner. It uses Docker/Singularity containers making installation trivial and results highly reproducible. The Nextflow DSL2 implementation of this pipeline uses one container per process which makes it much easier to maintain and update software dependencies. Where possible, these processes have been submitted to and installed from nf-core/modules in order to make them available to all nf-core pipelines, and to everyone within the Nextflow community!
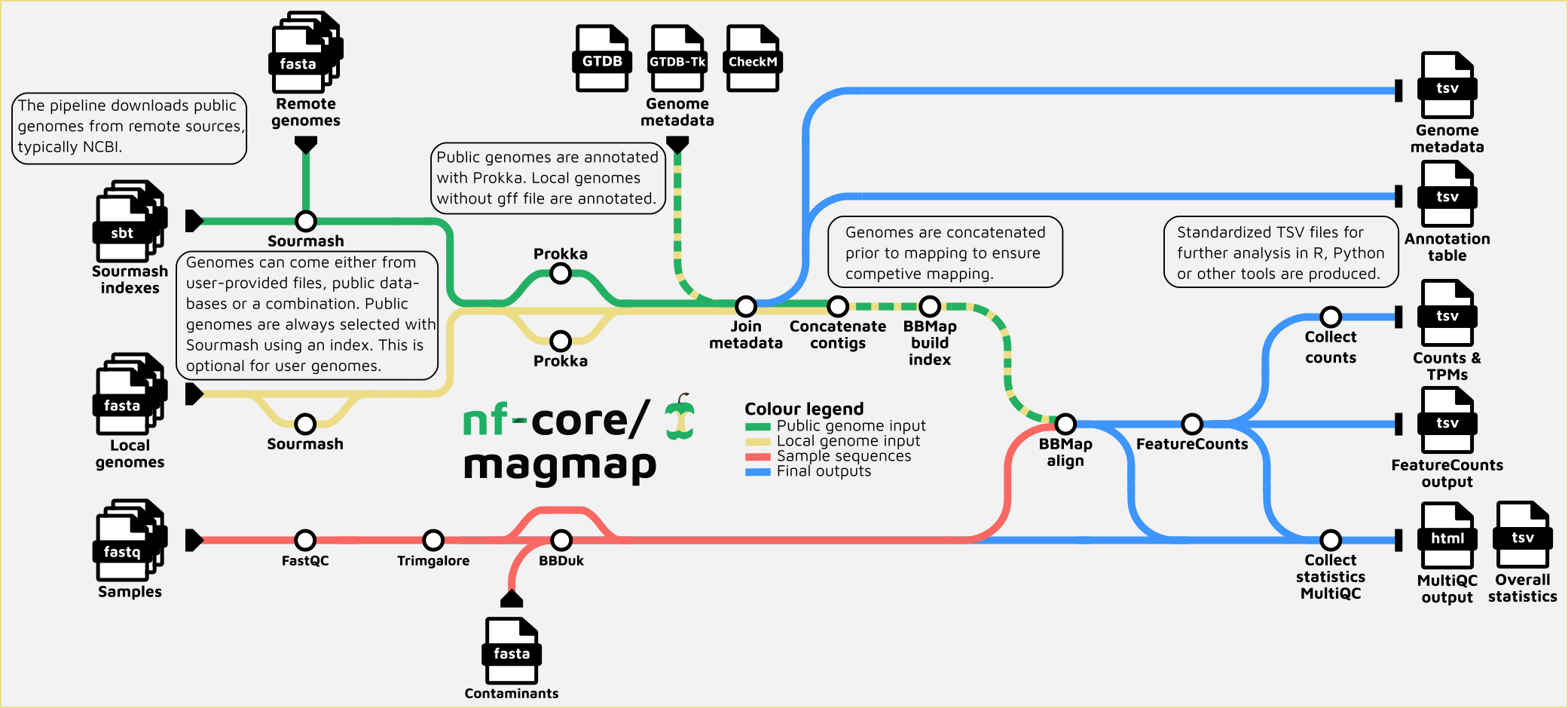
- Read QC (
FastQC) - Quality trimming and adapters removal for raw reads (
Trim Galore!) - Filter reads with
BBduk - Select reference genomes based on k-mer signatures in reads with
sourmash - Quantification of genes identified in selected reference genomes:
- Generate index of assembly (
BBmap index) - Mapping cleaned reads to the assembly for quantification (
BBmap) - Get raw counts per each gene present in the genomes (
Featurecounts) -> TSV table with collected featurecounts output
- Generate index of assembly (
- Summary statistics table. Collect_stats.R
- Overall summary of tools output, including QC for reads before and after trimming (
MultiQC)
Quick Start
If you are new to Nextflow and nf-core, please refer to this page on how to set-up Nextflow. Make sure to test your setup with -profile test before running the workflow on actual data.
First, prepare a samplesheet with your input data that looks as follows:
samplesheet.csv:
sample,fastq_1,fastq_2
CONTROL_REP1,AEG588A1_S1_L002_R1_001.fastq.gz,AEG588A1_S1_L002_R2_001.fastq.gz
CONTROL_REP2,AEG588A1_S2_L002_R1_001.fastq.gz,AEG588A1_S2_L002_R2_001.fastq.gzEach row represents a fastq file (single-end) or a pair of fastq files (paired end).
And, if you want to map to a set of your own genomes, a genome information file looking like this:
genomeinfo.csv:
accno,genome_fna,genome_gff
genome1,path/to/fna.gz,path/to/gff.gz
genome2,path/to/fna.gz,path/to/gff.gz
genome3,path/to/fna.gz,path/to/gff.gzEach row represents a genome file with or without the paired gff
Now, you can run the pipeline using:
nextflow run nf-core/magmap --input samplesheet.csv --genomeinfo genomeinfo.csv --outdir <OUTDIR> -profile <docker/singularity/podman/shifter/charliecloud/conda/institute>Please provide pipeline parameters via the CLI or Nextflow -params-file option. Custom config files including those provided by the -c Nextflow option can be used to provide any configuration except for parameters; see docs.
For more details and further functionality, please refer to the usage documentation and the parameter documentation.
Pipeline output
To see the results of an example test run with a full size dataset refer to the results tab on the nf-core website pipeline page. For more details about the output files and reports, please refer to the output documentation.
Credits
nf-core/magmap was originally written by Danilo Di Leo @danilodileo, Emelie Nilsson @emnillson and Daniel Lundin @erikrikardaniel.
Contributions and Support
If you would like to contribute to this pipeline, please see the contributing guidelines.
For further information or help, don’t hesitate to get in touch on the Slack #magmap channel (you can join with this invite).
Citations
An extensive list of references for the tools used by the pipeline can be found in the CITATIONS.md file.
You can cite the nf-core publication as follows:
The nf-core framework for community-curated bioinformatics pipelines.
Philip Ewels, Alexander Peltzer, Sven Fillinger, Harshil Patel, Johannes Alneberg, Andreas Wilm, Maxime Ulysse Garcia, Paolo Di Tommaso & Sven Nahnsen.
Nat Biotechnol. 2020 Feb 13. doi: 10.1038/s41587-020-0439-x.